Fluorescent Protein Guide: Empty Backbones荧光报告质粒载体 BioVector NTCC质粒载体菌种细胞基因保藏中心
- 价 格:¥19985
- 货 号:BioVector-Fluorescent Protein Guide: Empty Backbones
- 产 地:北京
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作QQ:1843439339 (微信同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
Background
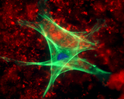
Fluorescent proteins are genetically encoded tools that are used extensively by life scientists. The original green fluorescent protein (GFP) was cloned in 1992 (Prasher et al., Gene, 1992), and since then scientists have engineered numerous GFP-variants and non-GFP proteins that result in a diverse set of colors.
BioVector has assembled a collection of empty plasmid backbones with different fluorescent tags for you to create fusion proteins with your gene of interest. These are organized By Color of the fluorescent protein and By Expression Species .
The content for this page was generated with the help of Erik Snapp .
Empty Backbones for Fluorescent Protein Fusions (Organized by Color)
For each table:
Excitation and Emission maximum are listed in nm.
Brightness is the product of exctinction coefficient and quantum yield divided by 1000.
pKa is the pH at which fluorescence intensity is 50% of its maximum value.
Maturation is the time for fluorescence to obtain half-maximal value.
Structure describes the monomeric or oligomeric state of the protein. Many FPs are monomers at low concentration but are prone to dimerization at high concentration, so this is noted here. Proteins that are derived from GFP and contain the A206K mutation are monomeric at all concentrations, so this is mutation is noted when present.
For more information on the properties of many of these proteins, see:
Cranfill et al., Nature Methods 2016 Jul;13(7):557-62.
FPbase: A database for users of fluorescent proteins created by Talley Lambert at Harvard Medical School
More References below
Jump to:
Blue/UV
Cyan
Green
Yellow
Orange
Red
Far-Red
Near-Infrared
Long Stokes Shift
Photoactivatable
Photoconvertible
Photoswitchable
Timers
Blue/UV
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| Sirius | 355 | 424 | 4 | 3.0 | Prone to dimerization |
| |
| SBFP2 | 380 | 446 | 13 | Monomer (A206K) |
| ||
| Azurite | 383 | 450 | 14 | 5 | Prone to dimerization |
| |
| mAzurite | 383 | 450 | 14 | 5 | Monomer (A206K) |
| |
| EBFP2 | 383 | 448 | 17 | 4.5 | Prone to dimerization |
| |
| moxBFP | 385 | 448 | 17 | Monomer (A206K) |
| ||
| mKalama1 | 385 | 456 | 16 | 5.5 | Monomer (A206K) |
| |
| mTagBFP2 | 399 | 456 | 32 | 2.7 | 12 min | Prone to dimerization |
|
Cyan
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| Aquamarine | 420 | 474 | 23 | 3.3 | Prone to dimerization |
| |
| ECFP | 433 | 475 | 13 | 4.7 | Prone to dimerization |
| |
| Cerulean | 433 | 475 | 27 | 4.7 | Monomer (A206K) |
| |
| mCerulean3 | 433 | 475 | 32 | 3.2 | Monomer (A206K) |
| |
| moxCerulean3 | 434 | 474 | 36 | 3.2 | Monomer (A206K) |
| |
| SCFP3A | 433 | 474 | 17 | 4.5 | Monomer (A206K) |
| |
| mTurquoise2 | 434 | 474 | 28 | 3.1 | Monomer (A206K) |
| |
| CyPet | 435 | 477 | 18 | 5 | Monomer |
| |
| AmCyan1 | 453 | 486 | 11 | Tetramer |
| ||
| MiCy (Midoriishi-Cyan) | 472 | 495 | 25 | 6.6 | Dimer |
|
Green
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| iLOV | 447 | 497 | Monomer |
| |||
| AcGFP1 | 475 | 505 | 28 | Prone to dimerization |
| ||
| sfGFP | 485 | 507 | 54 | Prone to dimerization |
| ||
| moxGFP | 486 | 510 | 50 | Monomer (A206K) |
| ||
| mEmerald | 487 | 509 | 39 | 6 | Monomer (A206K) |
| |
| EGFP | 488 | 507 | 34 | 6 | Prone to dimerization |
| |
| mEGFP | 488 | 507 | 34 | 6 | Monomer (A206K) |
| |
| mAzamiGreen | 492 | 505 | 41 | 5.8 | Monomer |
| |
| cfSGFP2 | 493 | 517 | 34 | 5.4 | Monomer (A206K) |
| |
| ZsGreen | 493 | 505 | 39 | Tetramer |
| ||
| SGFP2 | 495 | 512 | 30 | Monomer (A206K) |
| ||
| Clover | 505 | 515 | 84 | 6.1 | 30 min | Prone to dimerization |
|
| mClover2 | 505 | 515 | Monomer (A206K) |
| |||
| mClover3 | 506 | 518 | 85 | 6.5 | Monomer (A206K) |
| |
| mNeonGreen | 506 | 517 | 93 | 5.7 | 10 min | Monomer |
|
Yellow
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| EYFP | 513 | 527 | 51 | 6.9 | Prone to dimerization |
| |
| Topaz | 514 | 527 | 57 | Prone to dimerization |
| ||
| mTopaz | 514 | 527 | Monomer (A206K) |
| |||
| mVenus | 515 | 528 | 53 | 6 | Monomer (A206K) |
| |
| moxVenus | 514 | 526 | 44 | Monomer (A206K) |
| ||
| SYFP2 | 515 | 527 | 69 | 6 | Monomer (A206K) |
| |
| mGold | 515 | 530 | 68 | 5.9 | Monomer |
| |
| mCitrine | 516 | 529 | 59 | 5.7 | Monomer (A206K) |
| |
| YPet | 517 | 530 | 80 | 5.6 | Prone to dimerization |
| |
| ZsYellow1 | 529 | 539 | 8 | Tetramer |
| ||
| mPapaya1 | 530 | 541 | 36 | 6.8 | Monomer |
| |
| mCyRFP1 | 528 | 594 | 18 | 5.6 | Monomer |
|
Orange
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| mKusabira-Orange (mKO) | 548 | 559 | 31 | 5 | 4.5 hr | Monomer |
|
| mOrange | 548 | 562 | 49 | 6.5 | 2.5 hr | Monomer |
|
| mOrange2 | 549 | 565 | 35 | 6.5 | 4.5 hr | Monomer |
|
| mKO2 | 551 | 565 | 40 | 5.5 | Monomer |
| |
| TurboRFP | 553 | 574 | 62 | 4.4 | Dimer |
| |
| tdTomato | 554 | 581 | 95 | 4.7 | 1 hr | Tandem-dimer |
|
Red
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| mScarlet-H | 551 | 592 | 15 | 4.8 | 4.4 hr | Monomer |
|
| mNectarine | 558 | 578 | 26 | 6.9 | 30 min | Monomer |
|
| mRuby2 | 559 | 600 | 43 | 5.3 | 150 min | Prone to dimerization |
|
| eqFP611 | 559 | 611 | 35 | Tetramer |
| ||
| DsRed2 | 563 | 582 | 24 | Tetramer |
| ||
| mApple | 568 | 592 | 37 | 6.5 | 30 min | Monomer |
|
| mScarlet | 569 | 594 | 71 | 5.3 | 2.9 hr | Monomer |
|
| mScarlet-I | 569 | 593 | 57 | 5.4 | 0.6 hr | Monomer |
|
| mStrawberry | 574 | 596 | 26 | 4.5 | 50 min | Monomer |
|
| FusionRed | 580 | 608 | 18 | Monomer |
| ||
| mRFP1 | 584 | 607 | 13 | 4.5 | 1 hr | Monomer |
|
| mCherry | 587 | 610 | 16 | 4.5 | 15 min | Monomer |
|
| mCherry2 | 587 | 610 | Monomer |
|
Far-Red
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| mCrimson3 | 588 | 615 | - | Monomer |
| ||
| HcRed1 | 588 | 618 | 0.3 | Dimer |
| ||
| dKatushka | 588 | 635 | 22 | 5.5 | Dimer |
| |
| mKate1.3 | 588 | 635 | 25 | Monomer |
| ||
| mPlum | 590 | 649 | 4 | 4.5 | 100 min | Monomer |
|
| mRaspberry | 598 | 625 | 13 | 55 min | Monomer |
| |
| TagRFP675 | 598 | 675 | 4 | 5 | Prone to dimerization |
| |
| mNeptune | 600 | 650 | 13 | 5.4 | 35 min | Prone to dimerization |
|
| mNeptune2 | 600 | 650 | 21 | Prone to dimerization |
| ||
| mCarmine | 603 | 675 | 5.81 | 5.58 | Monomer |
| |
| mCardinal | 604 | 659 | 17 | 5.3 | 27 min | Prone to dimerization |
|
| mMaroon | 610 | 652 | - | Monomer |
| ||
| TagRFP657 | 611 | 657 | 0.34 | 5.3 | 27 min | Prone to dimerization |
|
| smURFP + BV | 642 | 670 | 32 | 39 min | Prone to dimerization |
|
Near-Infrared
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| miRFP670 | 642 | 670 | 12 | 4.5 | Monomer |
| |
| iRFP670 | 643 | 670 | 13 | 4 | ~5 hr | Dimer |
|
| iRFP682 | 663 | 682 | 10 | 4.5 | ~5hr | Dimer |
|
| miRFP703 | 673 | 703 | 8 | 4.5 | Monomer |
| |
| iRFP702 | 673 | 702 | 8 | 4.5 | ~5hr | Dimer |
|
| miRFP709 | 683 | 709 | 4 | 4.5 | Monomer |
| |
| mIFP | 683 | 704 | 7 | 3.5 | Monomer |
| |
| IFP2.0 | 684 | 708 | 7 | Prone to dimerization |
| ||
| iRFP (aka iRFP713) | 690 | 713 | 6 | 4.5 | 2.8 hr | Dimer |
|
| iSplit | 690 | 713 | 5 | Dimer |
| ||
| iRFP720 | 702 | 720 | 6 | 4.5 | ~5hr | Dimer |
|
Long Stokes Shift
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| T-sapphire | 399 | 511 | 26 | 4.9 | 1.3 hr | Prone to dimerization |
|
| mT-sapphire | 399 | 511 | Monomer (A206K) |
| |||
| mAmetrine | 406 | 526 | 26 | 6 | 0.8 hr | Monomer (A206K) |
|
| LSSmOrange | 437 | 572 | 23 | 5.7 | 2.3 hr | Monomer |
|
| mKeima Red | 440 | 620 | 3 | 6.5 | 4.4 hr | Monomer |
|
| dKeima Red | 440 | 616 | 22 | 6.5 | Dimer |
| |
| LSSmKate1 | 463 | 624 | 3 | 3.2 | 1.7 hr | Monomer |
|
| LSSmKate2 | 460 | 605 | 4 | 2.7 | 2.5 hr | Monomer |
|
Photoactivatable (e.g. off to on)
Excitation and Emission wavelengths after activation.
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| Phamret | 458 | 517 | 14 | Monomer |
| ||
| PA-sfGFP | 485 | 511 | Prone to dimerization |
| |||
| mPA-Emerald | 487 | 509 | - | Monomer (A206K) |
| ||
| PA-GFP | 504 | 517 | 14 | Monomer (A206K) |
| ||
| PATagRFP | 562 | 595 | 25 | 5.3 | 75 min | Prone to dimerization |
|
| PAmCherry1 | 564 | 595 | 8 | 6.3 | 23 min | Monomer |
|
| PAmCherry2 | 570 | 596 | 13 | 6.2 | 18 min | Monomer |
|
| PAmCherry3 | 570 | 596 | 5 | 6.2 | 25 min | Monomer |
|
| PAmKate | 568 | 628 | 5 | 5.6 | 19 min | Monomer |
|
| PAiRFP1 | 690 | 717 | 3 | Dimer |
| ||
| PAiRFP2 | 692 | 719 | 3 | Dimer |
|
Photoconvertible (e.g. green to red)
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| Dendra2 | 490 / 553 | 507 / 573 | 23/19 | 6.6/6.9 | Monomer |
| |
| mEos2 | 506 / 571 | 516 / 581 | 47 / 30 | 5.6/6.4 | Prone to oligomerization |
| |
| mEos3.2 | 507 / 572 | 516 / 580 | 53 / 18 | 5.4/5.8 | 20 min | Monomer |
|
| mEos4a | 505 / 569 | 516 / 581 | Prone to oligomerization |
| |||
| mEos4b | 505 / 569 | 516 / 581 | Monomer |
| |||
| dEos | 505 / 569 | 516 / 581 | 55 / 20 | Dimer |
| ||
| tdEos | 505 / 569 | 516 / 581 | 55 / 20 | Tandem dimer |
| ||
| mKikGR | 507 / 583 | 517 / 593 | 34 / 18 | 6.6/5.2 | Monomer |
| |
| Kaede | 508 / 572 | 518 / 580 | 87 / 20 | 5.6/5.6 | Tetramer |
| |
| PSmOrange | 548 / 634 | 565 / 662 | 58 / 9 | 6.2/5.6 | 1.6 hr | Monomer |
|
| PSmOrange2 | 546 / 619 | 561 / 651 | 31 / 7 | 6.6/5.4 | 3.5 hr | Monomer |
|
Photoswitchable (e.g. off to on to off)
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| rsTagRFP | 440 / 567 | 585 / 585 | 0.02 / 4.0 | 6.6 | 43 min | Prone to dimerization |
|
| rsEmerald | 491 | 508 |
| ||||
| rsGFP1 | 493 | 510 |
| ||||
| Dronpa3 | 503 | 518 | 19 | Monomer |
| ||
| mGeos-M | 503 / 569 | 514 / 581 | 44 | 4.7 | Prone to dimerization |
|
Fluorescent Timers
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Structure | Plasmids |
|---|---|---|---|---|---|---|---|
| Fast-FT | 403(blue), 583(red) | 466(blue), 606(red) | 14.9(blue), 6.8(red) | 2.8/4.1 | 7.1 hr (red) | Monomer |
|
| Medium-FT | 401(blue), 579(red) | 464(blue), 600(red) | 18.4(blue), 5.8(red) | 2.7/4.7 | 3.9 hr (red) | Monomer |
|
| Slow-FT | 402(blue), 583(red) | 465(blue), 604(red) | 11.7(blue), 4.2(red) | 2.6/4.6 | 28 hr (red) | Monomer |
|
- 用户
- 内容
- 询问日期
- 公告/新闻
- BioVector NTCC Inc.产品分类简介-下篇
- 2014-12-22
- BioVector NTCC Inc.产品分类中篇2
- 2014-12-22
- BioVector NTCC Inc.产品分类中篇1
- 2014-12-22
- BioVector NTCC产品分类目录简介-上篇
- 2014-12-22
- BioVector NTCC保藏中心简介
- 2014-11-20
- 人源/鼠源cDNA基因ORF克隆/表达质粒现货-BioVector NTCC保藏中心
- 2014-10-20
- 订购合同下载|汇款账户信息
- 2014-06-08


 免费订购电话: 400-800-2947
免费订购电话: 400-800-2947