大肠杆菌宿主菌HT115(DE3),RNase III缺陷型-BioVector NTCC质粒载体菌种细胞蛋白抗体基因保藏中心
- 价 格:¥9830
- 货 号:大肠杆菌宿主菌HT115(DE3)
- 产 地:北京
- BioVector NTCC典型培养物保藏中心
- 联系人:Dr.Xu, Biovector NTCC Inc.
电话:400-800-2947 工作微信:1843439339 (QQ同号)
邮件:Biovector@163.com
手机:18901268599
地址:北京
- 已注册
大肠杆菌宿主菌HT115(DE3),RNase III缺陷型
-BioVector NTCC质粒载体菌种细胞蛋白抗体基因保藏中心
Description
E. coli strain HT115(DE3) has a modified lac promotercontrolling the transcription of T7 RNA polymerase. It is an RNaseIII-deficient E. coli strain with IPTG-inducible T7 polymerase activity
This strain grows on LB or 2xYT plates (and is resistant totetracycline), and competent cells can be made using standard techniques.
Noteon tetracycline:
The Tn10 transposon interrupting the rnc14 gene carries atetracycline resistance gene. Therefore, bacteria should be subjected totetracycline selection (12.5 µg/ml) to maintain the RNase deficiency. However,the transposon is quite stable, as we have not lost it in the absence of selection.Using our protocol (see below and Kamathet al.Genome Biology,2, 1-10) inclusion oftetracycline during feeding significantly decreased the RNAi effect for severalgenes tested, so we recommend that it not be used in culture or in NGM plates duringfeeding using the method below. However, using the method of Timmons,etal.(Gene,263, 103-112), animprovement in feeding results by including tetracycline was reported.
NGMMedia:
For feeding plates, use standard NGM agar plus the followingingredients:
Carbenicillin to 25 µg/ml final concentration
IPTG to 1 mM final concentration
Plates are poured fresh 1-3 days before use.
Feeding Protocol(from Kamathet al. (2000) GenomeBiology, 2, 1-10):
1. Pick and grow bacteria 6 hours - overnight (but no longerthan 18 hours) in LB + 50 µg/ml ampicillin, seed onto NGM agar plates includingadditives (above). (Do not add IPTG or tetracycline to the liquid culture, asthis will reduce the RNAi effect.). Bacterial cultures grown shorter times (6hours) sometimes give better results. The lawn quality is improved if theculture is dried quickly by leaving the lids off for about 20 minutes afterseeding, but we don't know if this affects the RNAi effect. We also haveanecdotal evidence that plates poured at least 1 week before use work betterthan freshly poured plates.
2. Let dry and induce overnight at room temperature.
3. The following day, transfer an L4-stage hermaphrodite onto first plate,minimizing the amount of OP50 bacteria transferred (we usually wash worms in M9buffer and then aliquot them directly onto plates). Leave 72 hours at 15°C (or36-40 hours at 22°C) for RNAi to take effect, then replica plate adult ontoanother plate seeded with the same bacteria. After 24 hours, remove the adultfrom the replica and score the progeny for phenotypes. Alternatively, aliquotembryos or larvae onto the feeding plates and score them later.
4. Note on temperature:
We have observed that some genes give different phenotypes at 15°C vs 22°C;thus, it may be worthwhile to test a given gene using both conditions.
HT115 (DE3) genotype
F-, mcrA, mcrB, IN(rrnD-rrnE)1,lambda -, rnc14::Tn10(DE3 lysogen: lavUV5 promoter -T7 polymerase)(IPTG-inducible T7 polymerase) (RNase III minus).
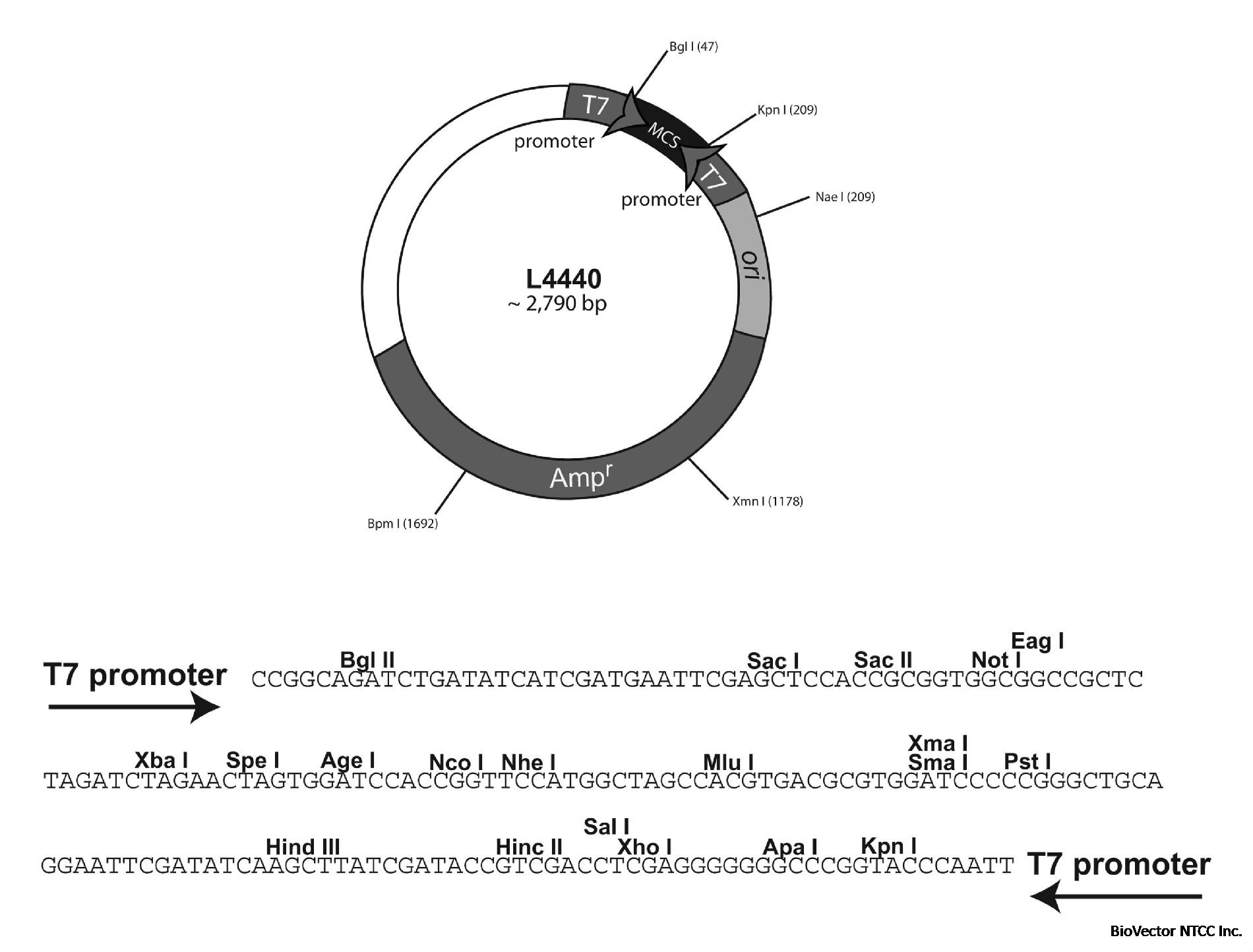
L4440线虫基因干扰载体RNAi质粒
T7启动子,Amp抗性,配套大肠杆菌宿主菌HT115(DE3)
BioVector NTCC质粒载体菌种细胞蛋白抗体基因保藏中心
- 公告/新闻